graphid.util package¶
Submodules¶
- graphid.util.mpl_plottool module
- graphid.util.mplutil module
multi_plot()figure()pandas_plot_matrix()axes_extent()extract_axes_extents()adjust_subplots()dict_intersection()_dark_background()_get_axis_xy_width_height()set_figtitle()legend()distinct_colors()distinct_markers()deterministic_shuffle()next_fnum()ensure_fnum()_save_requested()show_if_requested()save_parts()qtensure()imshow()colorbar()_get_plotdat()_set_plotdat()_del_plotdat()_get_plotdat_dict()_ensure_divider()scores_to_cmap()scores_to_color()reverse_colormap()PlotNumsdraw_border()draw_boxes()draw_line_segments()make_heatmask()Colorzoom_factory()pan_factory()PanEventsrelative_text()get_axis_xy_width_height()
- graphid.util.name_rectifier module
- graphid.util.nx_dynamic_graph module
GraphHelperMixinNiceGraphnx_UnionFindDynConnGraphDynConnGraph.clear()DynConnGraph.number_of_components()DynConnGraph.component()DynConnGraph.component_nodes()DynConnGraph.connected_to()DynConnGraph.node_label()DynConnGraph.node_labels()DynConnGraph.are_nodes_connected()DynConnGraph.connected_components()DynConnGraph.component_labels()DynConnGraph._cut()DynConnGraph._union()DynConnGraph._add_node()DynConnGraph._remove_node()DynConnGraph.add_edge()DynConnGraph.add_edges_from()DynConnGraph.add_node()DynConnGraph.add_nodes_from()DynConnGraph.remove_edge()DynConnGraph.remove_edges_from()DynConnGraph.remove_nodes_from()DynConnGraph.remove_node()DynConnGraph.subgraph()
- graphid.util.nx_utils module
_dz()nx_source_nodes()nx_sink_nodes()take_column()dict_take_column()itake_column()list_roll()diag_product()e_()edges_inside()edges_outgoing()edges_cross()edges_between()_edges_between_dense()_edges_inside_lower()_edges_inside_upper()_edges_between_disjoint()_edges_between_sparse()group_name_edges()ensure_multi_index()demodata_bridge()demodata_tarjan_bridge()is_k_edge_connected()complement_edges()k_edge_augmentation()is_complete()random_k_edge_connected_graph()edge_df()nx_delete_node_attr()nx_delete_edge_attr()nx_gen_node_values()nx_gen_node_attrs()graph_info()assert_raises()bfs_conditional()nx_gen_edge_attrs()nx_gen_edge_values()nx_edges()nx_delete_None_edge_attr()nx_delete_None_node_attr()nx_node_dict()
- graphid.util.priority_queue module
- graphid.util.util_boxes module
- graphid.util.util_grabdata module
- graphid.util.util_graphviz module
dump_nx_ondisk()ensure_nonhex_color()show_nx()netx_draw_images_at_positions()parse_html_graphviz_attrs()GRAPHVIZ_KEYSget_explicit_graph()get_nx_layout()apply_graph_layout_attrs()patch_pygraphviz()make_agraph()_groupby_prelayout()nx_agraph_layout()parse_point()parse_anode_layout_attrs()parse_aedge_layout_attrs()_get_node_size()draw_network2()stack_graphs()translate_graph()translate_graph_to_origin()get_graph_bounding_box()nx_ensure_agraph_color()bbox_from_extent()get_pointset_extents()
- graphid.util.util_group module
- graphid.util.util_image module
- graphid.util.util_kw module
- graphid.util.util_misc module
randn()aslist()classpropertyestarmap()delete_dict_keys()flag_None_items()where()delete_items_by_index()make_index_lookup()cprint()ensure_iterable()highlight_regex()regex_word()setdiff()all_dict_combinations()iteritems_sorted()partial_order()replace_nones()take_percentile_parts()snapped_slice()get_timestamp()isect()safe_extreme()safe_argmax()safe_max()safe_min()stats_dict()
- graphid.util.util_numpy module
- graphid.util.util_random module
- graphid.util.util_tags module
Module contents¶
mkinit graphid.util
- class graphid.util.Color(color, alpha=None, space=None)[source]¶
Bases:
NiceReprmove to colorutil?
Example
>>> print(Color('g')) >>> print(Color('orangered')) >>> print(Color('#AAAAAA').as255()) >>> print(Color([0, 255, 0])) >>> print(Color([1, 1, 1.])) >>> print(Color([1, 1, 1])) >>> print(Color(Color([1, 1, 1])).as255()) >>> print(Color(Color([1., 0, 1, 0])).ashex()) >>> print(Color([1, 1, 1], alpha=255)) >>> print(Color([1, 1, 1], alpha=255, space='lab'))
- classmethod _is_base255(channels)[source]¶
there is a one corner case where all pixels are 1 or less
- adjust_hsv(hue_adjust=0.0, sat_adjust=0.0, val_adjust=0.0)[source]¶
Performs adaptive changes to the HSV values of the color.
- Parameters:
hue_adjust (float) – addative
sat_adjust (float)
val_adjust (float)
- Returns:
new_rgb
- Return type:
CommandLine
python -m graphid.util.mplutil Color.adjust_hsv
Example
>>> rgb_list = [Color(c).as01() for c in ['pink', 'yellow', 'green']] >>> hue_adjust = -0.1 >>> sat_adjust = +0.5 >>> val_adjust = -0.1 >>> # execute function >>> new_rgb_list = [Color(rgb).adjust_hsv(hue_adjust, sat_adjust, val_adjust) for rgb in rgb_list] >>> print(ub.urepr(new_rgb_list, nl=1, sv=True)) [ <Color(rgb: 0.90, 0.23, 0.75)>, <Color(rgb: 0.90, 0.36, 0.00)>, <Color(rgb: 0.24, 0.40, 0.00)>, ] >>> # xdoc: +REQUIRES(--show) >>> color_list = rgb_list + new_rgb_list >>> testshow_colors(color_list)
- class graphid.util.PlotNums(nRows=None, nCols=None, nSubplots=None, start=0)[source]¶
Bases:
objectConvinience class for dealing with plot numberings (pnums)
Example
>>> pnum_ = PlotNums(nRows=2, nCols=2) >>> # Indexable >>> print(pnum_[0]) (2, 2, 1) >>> # Iterable >>> print(ub.urepr(list(pnum_), nl=0, nobr=1)) (2, 2, 1), (2, 2, 2), (2, 2, 3), (2, 2, 4) >>> # Callable (iterates through a default iterator) >>> print(pnum_()) (2, 2, 1) >>> print(pnum_()) (2, 2, 2)
- classmethod _get_num_rc(nSubplots=None, nRows=None, nCols=None)[source]¶
Gets a constrained row column plot grid
- Parameters:
nSubplots (None) – (default = None)
nRows (None) – (default = None)
nCols (None) – (default = None)
- Returns:
(nRows, nCols)
- Return type:
Example
>>> cases = [ >>> dict(nRows=None, nCols=None, nSubplots=None), >>> dict(nRows=2, nCols=None, nSubplots=5), >>> dict(nRows=None, nCols=2, nSubplots=5), >>> dict(nRows=None, nCols=None, nSubplots=5), >>> ] >>> for kw in cases: >>> print('----') >>> size = PlotNums._get_num_rc(**kw) >>> if kw['nSubplots'] is not None: >>> assert size[0] * size[1] >= kw['nSubplots'] >>> print('**kw = %s' % (ub.urepr(kw),)) >>> print('size = %r' % (size,))
- _get_square_row_cols(max_cols=None, fix=False, inclusive=True)[source]¶
- Parameters:
nSubplots (int)
max_cols (int)
- Returns:
(int, int)
- Return type:
Example
>>> nSubplots = 9 >>> nSubplots_list = [1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11] >>> max_cols = None >>> rc_list = [PlotNums._get_square_row_cols(nSubplots, fix=True) for nSubplots in nSubplots_list] >>> print(repr(np.array(rc_list).T)) array([[1, 1, 2, 2, 2, 2, 3, 3, 3, 3, 3], [1, 2, 2, 2, 3, 3, 3, 3, 3, 4, 4]])
- graphid.util.adjust_subplots(left=None, right=None, bottom=None, top=None, wspace=None, hspace=None, fig=None)[source]¶
- Kwargs:
left (float): left side of the subplots of the figure right (float): right side of the subplots of the figure bottom (float): bottom of the subplots of the figure top (float): top of the subplots of the figure wspace (float): width reserved for blank space between subplots hspace (float): height reserved for blank space between subplots
- graphid.util.axes_extent(axs, pad=0.0)[source]¶
Get the full extent of a group of axes, including axes labels, tick labels, and titles.
- graphid.util.colorbar(scalars, colors, custom=False, lbl=None, ticklabels=None, float_format='%.2f', **kwargs)[source]¶
adds a color bar next to the axes based on specific scalars
- Parameters:
scalars (ndarray)
colors (ndarray)
custom (bool) – use custom ticks
- Kwargs:
See plt.colorbar
- Returns:
matplotlib colorbar object
- Return type:
cb
- graphid.util.deterministic_shuffle(list_, rng=0)[source]¶
-
Example
>>> list_ = [1, 2, 3, 4, 5, 6] >>> seed = 1 >>> list_ = deterministic_shuffle(list_, seed) >>> result = str(list_) >>> print(result) [3, 2, 5, 1, 4, 6]
- graphid.util.dict_intersection(dict1, dict2)[source]¶
Key AND Value based dictionary intersection
- Parameters:
dict1 (dict)
dict2 (dict)
- Returns:
- Return type:
Example
>>> dict1 = {'a': 1, 'b': 2, 'c': 3, 'd': 4} >>> dict2 = {'b': 2, 'c': 3, 'd': 5, 'e': 21, 'f': 42} >>> mergedict_ = dict_intersection(dict1, dict2) >>> print(ub.urepr(mergedict_, nl=0, sort=1)) {'b': 2, 'c': 3}
- graphid.util.distinct_colors(N, brightness=0.878, randomize=True, hue_range=(0.0, 1.0), cmap_seed=None)[source]¶
- Parameters:
N (int)
brightness (float)
- Returns:
RGB_tuples
- Return type:
CommandLine
python -m color_funcs --test-distinct_colors --N 2 --show --hue-range=0.05,.95 python -m color_funcs --test-distinct_colors --N 3 --show --hue-range=0.05,.95 python -m color_funcs --test-distinct_colors --N 4 --show --hue-range=0.05,.95 python -m .color_funcs --test-distinct_colors --N 3 --show --no-randomize python -m .color_funcs --test-distinct_colors --N 4 --show --no-randomize python -m .color_funcs --test-distinct_colors --N 6 --show --no-randomize python -m .color_funcs --test-distinct_colors --N 20 --show
References
http://blog.jianhuashao.com/2011/09/generate-n-distinct-colors.html
- graphid.util.distinct_markers(num, style='astrisk', total=None, offset=0)[source]¶
- Parameters:
num (?)
- graphid.util.draw_border(ax, color, lw=2, offset=None, adjust=True)[source]¶
draws rectangle border around a subplot
- graphid.util.draw_boxes(boxes, box_format='xywh', color='blue', labels=None, textkw=None, ax=None)[source]¶
- Parameters:
boxes (list) – list of coordindates in xywh, tlbr, or cxywh format
box_format (str) – specify how boxes are formated xywh is the top left x and y pixel width and height cxywh is the center xy pixel width and height tlbr is the top left xy and the bottom right xy
color (str) – edge color of the boxes
labels (list) – if specified, plots a text annotation on each box
Example
>>> qtensure() # xdoc: +SKIP >>> bboxes = [[.1, .1, .6, .3], [.3, .5, .5, .6]] >>> col = draw_boxes(bboxes)
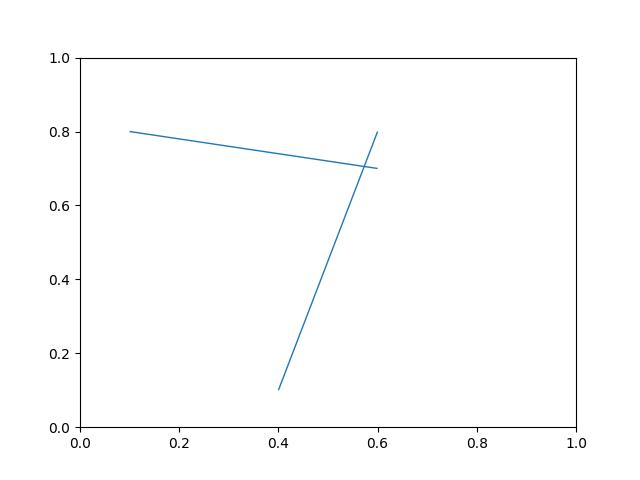
- graphid.util.draw_line_segments(pts1, pts2, ax=None, **kwargs)[source]¶
draws N line segments between N pairs of points
- Parameters:
pts1 (ndarray) – Nx2
pts2 (ndarray) – Nx2
ax (None) – (default = None)
**kwargs – lw, alpha, colors
Example
>>> pts1 = np.array([(.1, .8), (.6, .8)]) >>> pts2 = np.array([(.6, .7), (.4, .1)]) >>> figure(fnum=None) >>> draw_line_segments(pts1, pts2) >>> # xdoc: +REQUIRES(--show) >>> import matplotlib.pyplot as plt >>> ax = plt.gca() >>> ax.set_xlim(0, 1) >>> ax.set_ylim(0, 1) >>> show_if_requested()
- graphid.util.figure(fnum=None, pnum=(1, 1, 1), title=None, figtitle=None, doclf=False, docla=False, projection=None, **kwargs)[source]¶
http://matplotlib.org/users/gridspec.html
- Parameters:
fnum (int) – fignum = figure number
pnum (int, str, or tuple(int, int, int)) – plotnum = plot tuple
title (str) – (default = None)
figtitle (None) – (default = None)
docla (bool) – (default = False)
doclf (bool) – (default = False)
- Returns:
fig
- Return type:
mpl.Figure
Example
>>> import matplotlib.pyplot as plt >>> fnum = 1 >>> fig = figure(fnum, (2, 2, 1)) >>> plt.gca().text(0.5, 0.5, "ax1", va="center", ha="center") >>> fig = figure(fnum, (2, 2, 2)) >>> plt.gca().text(0.5, 0.5, "ax2", va="center", ha="center") >>> show_if_requested()
Example
>>> import matplotlib.pyplot as plt >>> fnum = 1 >>> fig = figure(fnum, (2, 2, 1)) >>> plt.gca().text(0.5, 0.5, "ax1", va="center", ha="center") >>> fig = figure(fnum, (2, 2, 2)) >>> plt.gca().text(0.5, 0.5, "ax2", va="center", ha="center") >>> fig = figure(fnum, (2, 4, (1, slice(1, None)))) >>> plt.gca().text(0.5, 0.5, "ax3", va="center", ha="center") >>> show_if_requested()
- graphid.util.get_axis_xy_width_height(ax=None, xaug=0, yaug=0, waug=0, haug=0)[source]¶
gets geometry of a subplot
- graphid.util.imshow(img, fnum=None, title=None, figtitle=None, pnum=None, interpolation='nearest', cmap=None, heatmap=False, data_colorbar=False, xlabel=None, redraw_image=True, colorspace='bgr', ax=None, alpha=None, norm=None, **kwargs)[source]¶
- Parameters:
img (ndarray) – image data
fnum (int) – figure number
colorspace (str) – if the data is 3-4 channels, this indicates the colorspace 1 channel data is assumed grayscale. 4 channels assumes alpha.
title (str)
figtitle (None)
pnum (tuple) – plot number
interpolation (str) – other interpolations = nearest, bicubic, bilinear
cmap (None)
heatmap (bool)
data_colorbar (bool)
darken (None)
redraw_image (bool) – used when calling imshow over and over. if false doesnt do the image part.
- Returns:
(fig, ax)
- Return type:
- Kwargs:
docla, doclf, projection
- Returns:
(fig, ax)
- Return type:
- graphid.util.legend(loc='best', fontproperties=None, size=None, fc='w', alpha=1, ax=None, handles=None)[source]¶
- Parameters:
loc (str) – (default = ‘best’)
fontproperties (None) – (default = None)
size (None) – (default = None)
- graphid.util.make_heatmask(probs, cmap='plasma', with_alpha=True)[source]¶
Colorizes a single-channel intensity mask (with an alpha channel)
- graphid.util.multi_plot(xdata=None, ydata=[], **kwargs)[source]¶
plots multiple lines, bars, etc…
This is the big function that implements almost all of the heavy lifting in this file. Any function not using this should probably find a way to use it. It is pretty general and relatively clean.
- Parameters:
xdata (ndarray) – can also be a list of arrays
ydata (list or dict of ndarrays) – can also be a single array
**kwargs –
- Misc:
fnum, pnum, use_legend, legend_loc
- Labels:
xlabel, ylabel, title, figtitle ticksize, titlesize, legendsize, labelsize
- Grid:
gridlinewidth, gridlinestyle
- Ticks:
num_xticks, num_yticks, tickwidth, ticklength, ticksize
- Data:
xmin, xmax, ymin, ymax, spread_list # can append _list to any of these # these can be dictionaries if ydata was also a dict
- plot_kw_keys = [‘label’, ‘color’, ‘marker’, ‘markersize’,
‘markeredgewidth’, ‘linewidth’, ‘linestyle’]
any plot_kw key can be a scalar (corresponding to all ydatas), a list if ydata was specified as a list, or a dict if ydata was specified as a dict.
kind = [‘bar’, ‘plot’, …]
- if kind=’plot’:
spread
- if kind=’bar’:
stacked, width
References
matplotlib.org/examples/api/barchart_demo.html
Example
>>> xdata = [1, 2, 3, 4, 5] >>> ydata_list = [[1, 2, 3, 4, 5], [3, 3, 3, 3, 3], [5, 4, np.nan, 2, 1], [4, 3, np.nan, 1, 0]] >>> kwargs = {'label': ['spamΣ', 'eggs', 'jamµ', 'pram'], 'linestyle': '-'} >>> #fig = multi_plot(xdata, ydata_list, title='$\phi_1(\\vec{x})$', xlabel='\nfds', **kwargs) >>> fig = multi_plot(xdata, ydata_list, title='ΣΣΣµµµ', xlabel='\nfdsΣΣΣµµµ', **kwargs) >>> show_if_requested()
Example
>>> fig1 = multi_plot([1, 2, 3], [4, 5, 6]) >>> fig2 = multi_plot([1, 2, 3], [4, 5, 6], fnum=4) >>> show_if_requested()
- graphid.util.pandas_plot_matrix(df, rot=90, ax=None, grid=True, label=None, zerodiag=False, cmap='viridis', showvals=False, logscale=True)[source]¶
- graphid.util.relative_text(pos, text, ax=None, offset=None, **kwargs)[source]¶
Places text on axes in a relative position
- Parameters:
pos (tuple) – relative xy position
text (str) – text
ax (None) – (default = None)
offset (None) – (default = None)
**kwargs – horizontalalignment, verticalalignment, roffset, ha, va, fontsize, fontproperties, fontproperties, clip_on
CommandLine
python -m graphid.util.mplutil relative_text --show
Example
>>> from graphid import util >>> import matplotlib as mpl >>> x = .5 >>> y = .5 >>> util.figure() >>> txt = 'Hello World' >>> family = 'monospace' >>> family = 'CMU Typewriter Text' >>> fontproperties = mpl.font_manager.FontProperties(family=family, >>> size=42) >>> relative_text((x, y), txt, halign='center', >>> fontproperties=fontproperties) >>> util.show_if_requested()
- graphid.util.reverse_colormap(cmap)[source]¶
References
http://nbviewer.ipython.org/github/kwinkunks/notebooks/blob/master/Matteo_colourmaps.ipynb
- graphid.util.save_parts(fig, fpath, grouped_axes=None, dpi=None)[source]¶
FIXME: this works in mpl 2.0.0, but not 2.0.2
- Parameters:
fig (?)
fpath (str) – file path string
dpi (None) – (default = None)
- Returns:
subpaths
- Return type:
CommandLine
python -m draw_func2 save_parts
- graphid.util.scores_to_color(score_list, cmap_='hot', logscale=False, reverse_cmap=False, custom=False, val2_customcolor=None, score_range=None, cmap_range=(0.1, 0.9))[source]¶
Other good colormaps are ‘spectral’, ‘gist_rainbow’, ‘gist_ncar’, ‘Set1’, ‘Set2’, ‘Accent’ # TODO: plasma
- Parameters:
score_list (list)
cmap_ (str) – defaults to hot
logscale (bool)
cmap_range (tuple) – restricts to only a portion of the cmap to avoid extremes
- Returns:
<class ‘_ast.ListComp’>
- graphid.util.set_figtitle(figtitle, subtitle='', forcefignum=True, incanvas=True, size=None, fontfamily=None, fontweight=None, fig=None)[source]¶
- Parameters:
figtitle (?)
subtitle (str) – (default = ‘’)
forcefignum (bool) – (default = True)
incanvas (bool) – (default = True)
fontfamily (None) – (default = None)
fontweight (None) – (default = None)
size (None) – (default = None)
fig (None) – (default = None)
CommandLine
python -m .custom_figure set_figtitle --show
Example
>>> # DISABLE_DOCTEST >>> fig = figure(fnum=1, doclf=True) >>> result = set_figtitle(figtitle='figtitle', fig=fig) >>> # xdoc: +REQUIRES(--show) >>> show_if_requested()
- graphid.util.show_if_requested(N=1)[source]¶
Used at the end of tests. Handles command line arguments for saving figures
- graphid.util.zoom_factory(ax=None, zoomable_list=[], base_scale=1.1)[source]¶
References
https://gist.github.com/tacaswell/3144287 http://stackoverflow.com/questions/11551049/matplotlib-plot-zooming-with-scroll-wheel
- graphid.util.demodata_oldnames(n_incon_groups=10, n_con_groups=2, n_per_con=5, n_per_incon=5, con_sep=4, n_empty_groups=0)[source]¶
- graphid.util.find_consistent_labeling(grouped_oldnames, extra_prefix='_extra_name', verbose=False)[source]¶
Solves a a maximum bipirtite matching problem to find a consistent name assignment that minimizes the number of annotations with different names. For each new grouping of annotations we assign
For each group of annotations we must assign them all the same name, either from
To reduce the running time
- Parameters:
gropued_oldnames (list) – A group of old names where the grouping is based on new names. For instance:
- Given:
aids = [1, 2, 3, 4, 5] old_names = [0, 1, 1, 1, 0] new_names = [0, 0, 1, 1, 0]
- The grouping is
[[0, 1, 0], [1, 1]]
This lets us keep the old names in a split case and re-use exising names and make minimal changes to current annotation names while still being consistent with the new and improved grouping.
- The output will be:
[0, 1]
Meaning that all annots in the first group are assigned the name 0 and all annots in the second group are assigned the name 1.
References
http://stackoverflow.com/questions/1398822/assignment-problem-numpy
Example
>>> grouped_oldnames = demodata_oldnames(25, 15, 5, n_per_incon=5) >>> new_names = find_consistent_labeling(grouped_oldnames, verbose=1) >>> grouped_oldnames = demodata_oldnames(0, 15, 5, n_per_incon=1) >>> new_names = find_consistent_labeling(grouped_oldnames, verbose=1) >>> grouped_oldnames = demodata_oldnames(0, 0, 0, n_per_incon=1) >>> new_names = find_consistent_labeling(grouped_oldnames, verbose=1)
Example
>>> # xdoctest: +REQUIRES(module:timerit) >>> import timerit >>> ydata = [] >>> xdata = list(range(10, 150, 50)) >>> for x in xdata: >>> print('x = %r' % (x,)) >>> grouped_oldnames = demodata_oldnames(x, 15, 5, n_per_incon=5) >>> t = timerit.Timerit(3, verbose=1) >>> for timer in t: >>> with timer: >>> new_names = find_consistent_labeling(grouped_oldnames) >>> ydata.append(t.min()) >>> # xdoc: +REQUIRES(--show) >>> import plottool_ibeis as pt >>> pt.qtensure() >>> pt.multi_plot(xdata, [ydata]) >>> util.show_if_requested()
Example
>>> grouped_oldnames = [['a', 'b', 'c'], ['b', 'c'], ['c', 'e', 'e']] >>> new_names = find_consistent_labeling(grouped_oldnames, verbose=1) >>> result = ub.urepr(new_names) >>> print(new_names) ['a', 'b', 'e']
Example
>>> grouped_oldnames = [['a', 'b'], ['a', 'a', 'b'], ['a']] >>> new_names = find_consistent_labeling(grouped_oldnames) >>> result = ub.urepr(new_names) >>> print(new_names) ['b', 'a', '_extra_name0']
Example
>>> grouped_oldnames = [['a', 'b'], ['e'], ['a', 'a', 'b'], [], ['a'], ['d']] >>> new_names = find_consistent_labeling(grouped_oldnames) >>> result = ub.urepr(new_names) >>> print(new_names) ['b', 'e', 'a', '_extra_name0', '_extra_name1', 'd']
Example
>>> grouped_oldnames = [[], ['a', 'a'], [], >>> ['a', 'a', 'a', 'a', 'a', 'a', 'a', 'b'], ['a']] >>> new_names = find_consistent_labeling(grouped_oldnames) >>> result = ub.urepr(new_names) >>> print(new_names) ['_extra_name0', 'a', '_extra_name1', 'b', '_extra_name2']
- graphid.util.simple_munkres(part_oldnames)[source]¶
Defines a munkres problem to solve name rectification.
Notes
We create a matrix where each rows represents a group of annotations in the same PCC and each column represents an original name. If there are more PCCs than original names the columns are padded with extra values. The matrix is first initialized to be negative infinity representing impossible assignments. Then for each column representing a padded name, we set we its value to $1$ indicating that each new name could be assigned to a padded name for some small profit. Finally, let $f_{rc}$ be the the number of annotations in row $r$ with an original name of $c$. Each matrix value $(r, c)$ is set to $f_{rc} + 1$ if $f_{rc} > 0$, to represent how much each name ``wants’’ to be labeled with a particular original name, and the extra one ensures that these original names are always preferred over padded names.
Example
>>> part_oldnames = [['a', 'b'], ['b', 'c'], ['c', 'a', 'a']] >>> new_names = simple_munkres(part_oldnames) >>> result = ub.urepr(new_names) >>> print(new_names) ['b', 'c', 'a']
Example
>>> part_oldnames = [[], ['a', 'a'], [], >>> ['a', 'a', 'a', 'a', 'a', 'a', 'a', 'b'], ['a']] >>> new_names = simple_munkres(part_oldnames) >>> result = ub.urepr(new_names) >>> print(new_names) [None, 'a', None, 'b', None]
Example
>>> part_oldnames = [[], ['b'], ['a', 'b', 'c'], ['b', 'c'], ['c', 'e', 'e']] >>> new_names = find_consistent_labeling(part_oldnames) >>> result = ub.urepr(new_names) >>> print(new_names) ['_extra_name0', 'b', 'a', 'c', 'e']
- Profit Matrix
b a c e _0
0 -10 -10 -10 -10 1 1 2 -10 -10 -10 1 2 2 2 2 -10 1 3 2 -10 2 -10 1 4 -10 -10 2 3 1
- class graphid.util.DynConnGraph(*args, **kwargs)[source]¶
Bases:
Graph,GraphHelperMixinDynamically connected graph.
Maintains a data structure parallel to a normal networkx graph that maintains dynamic connectivity for fast connected compoment queries.
Underlying Data Structures and limitations are
Data Structure | Insertion | Deletion | CC Find |¶
UnionFind | lg(n) | n | No UnionFind2 | n* | n | 1 EulerTourForest | lg^2(n) | lg^2(n) | lg(n) / lglg(n) - - Ammortized
O(n) is worst case, but it seems to be very quick in practice. The average runtime should be close to lg(n), but I’m writing this doc 8 months after working on this algo, so I may not remember exactly.
References
https://courses.csail.mit.edu/6.851/spring14/lectures/L20.pdf https://courses.csail.mit.edu/6.851/spring14/lectures/L20.html http://cs.stackexchange.com/questions/33595/maintaining-connecte https://en.wikipedia.org/wiki/Dynamic_connectivity#Fully_dynamic_connectivity
Example
>>> self = DynConnGraph() >>> self.add_edges_from([(1, 2), (2, 3), (4, 5), (6, 7), (7, 4)]) >>> self.add_edges_from([(10, 20), (20, 30), (40, 50), (60, 70), (70, 40)]) >>> self._ccs >>> u, v = 20, 1 >>> assert self.node_label(u) != self.node_label(v) >>> assert self.connected_to(u) != self.connected_to(v) >>> self.add_edge(u, v) >>> assert self.node_label(u) == self.node_label(v) >>> assert self.connected_to(u) == self.connected_to(v) >>> self.remove_edge(u, v) >>> assert self.node_label(u) != self.node_label(v) >>> assert self.connected_to(u) != self.connected_to(v) >>> ccs = list(self.connected_components()) >>> # xdoctest: +REQUIRES(--show) >>> import plottool_ibeis as pt >>> pt.qtensure() >>> pt.show_nx(self)
# todo: check if nodes exist when adding
- component_nodes(label)¶
- node_label(node)[source]¶
Example
>>> self = DynConnGraph() >>> self.add_edges_from([(1, 2), (2, 3), (4, 5), (6, 7)]) >>> assert self.node_label(2) == self.node_label(1) >>> assert self.node_label(2) != self.node_label(4)
- connected_components()[source]¶
Example
>>> self = DynConnGraph() >>> self.add_edges_from([(1, 2), (2, 3), (4, 5), (6, 7)]) >>> ccs = list(self.connected_components()) >>> result = 'ccs = {}'.format(ub.urepr(ccs, nl=0)) >>> print(result) ccs = [{1, 2, 3}, {4, 5}, {6, 7}]
- add_edge(u, v, **attr)[source]¶
Example
>>> self = DynConnGraph() >>> self.add_edges_from([(1, 2), (2, 3), (4, 5), (6, 7), (7, 4)]) >>> assert self._ccs == {1: {1, 2, 3}, 4: {4, 5, 6, 7}} >>> self.add_edge(1, 5) >>> assert self._ccs == {1: {1, 2, 3, 4, 5, 6, 7}}
- remove_edge(u, v)[source]¶
Example
>>> self = DynConnGraph() >>> self.add_edges_from([(1, 2), (2, 3), (4, 5), (6, 7), (7, 4)]) >>> assert self._ccs == {1: {1, 2, 3}, 4: {4, 5, 6, 7}} >>> self.add_edge(1, 5) >>> assert self._ccs == {1: {1, 2, 3, 4, 5, 6, 7}} >>> self.remove_edge(1, 5) >>> assert self._ccs == {1: {1, 2, 3}, 4: {4, 5, 6, 7}}
- remove_node(n)[source]¶
Example
>>> self = DynConnGraph() >>> self.add_edges_from([(1, 2), (2, 3), (4, 5), (5, 6), (6, 7), (7, 8), (8, 9)]) >>> assert self._ccs == {1: {1, 2, 3}, 4: {4, 5, 6, 7, 8, 9}} >>> self.remove_node(2) >>> assert self._ccs == {1: {1}, 3: {3}, 4: {4, 5, 6, 7, 8, 9}} >>> self.remove_node(7) >>> assert self._ccs == {1: {1}, 3: {3}, 4: {4, 5, 6}, 8: {8, 9}}
- class graphid.util.GraphHelperMixin[source]¶
Bases:
NiceReprEnsures that we always return edges in a consistent order
- class graphid.util.NiceGraph(incoming_graph_data=None, **attr)[source]¶
Bases:
Graph,GraphHelperMixinInitialize a graph with edges, name, or graph attributes.
- Parameters:
incoming_graph_data (input graph (optional, default: None)) – Data to initialize graph. If None (default) an empty graph is created. The data can be an edge list, or any NetworkX graph object. If the corresponding optional Python packages are installed the data can also be a 2D NumPy array, a SciPy sparse array, or a PyGraphviz graph.
attr (keyword arguments, optional (default= no attributes)) – Attributes to add to graph as key=value pairs.
See also
convertExamples
>>> G = nx.Graph() # or DiGraph, MultiGraph, MultiDiGraph, etc >>> G = nx.Graph(name="my graph") >>> e = [(1, 2), (2, 3), (3, 4)] # list of edges >>> G = nx.Graph(e)
Arbitrary graph attribute pairs (key=value) may be assigned
>>> G = nx.Graph(e, day="Friday") >>> G.graph {'day': 'Friday'}
- graphid.util.assert_raises(ex_type, func, *args, **kwargs)[source]¶
Checks that a function raises an error when given specific arguments.
- Parameters:
ex_type (Exception) – exception type
func (callable) – live python function
Example
>>> ex_type = AssertionError >>> func = len >>> assert_raises(ex_type, assert_raises, ex_type, func, []) >>> assert_raises(ValueError, [].index, 0)
- graphid.util.bfs_conditional(G, source, reverse=False, keys=True, data=False, yield_nodes=True, yield_if=None, continue_if=None, visited_nodes=None, yield_source=False)[source]¶
Produce edges in a breadth-first-search starting at source, but only return nodes that satisfiy a condition, and only iterate past a node if it satisfies a different condition.
conditions are callables that take (G, child, edge) and return true or false
Example
>>> import networkx as nx >>> G = nx.Graph() >>> G.add_edges_from([(1, 2), (1, 3), (2, 3), (2, 4)]) >>> continue_if = lambda G, child, edge: True >>> result = list(bfs_conditional(G, 1, yield_nodes=False)) >>> print(result) [(1, 2), (1, 3), (2, 1), (2, 3), (2, 4), (3, 1), (3, 2), (4, 2)]
Example
>>> import networkx as nx >>> G = nx.Graph() >>> continue_if = lambda G, child, edge: (child % 2 == 0) >>> yield_if = lambda G, child, edge: (child % 2 == 1) >>> G.add_edges_from([(0, 1), (1, 3), (3, 5), (5, 10), >>> (4, 3), (3, 6), >>> (0, 2), (2, 4), (4, 6), (6, 10)]) >>> result = list(bfs_conditional(G, 0, continue_if=continue_if, >>> yield_if=yield_if)) >>> print(result) [1, 3, 5]
- graphid.util.demodata_tarjan_bridge()[source]¶
Example
>>> from graphid import util >>> G = demodata_tarjan_bridge() >>> # xdoc: +REQUIRES(--show) >>> util.show_nx(G) >>> util.show_if_requested()
- graphid.util.edges_between(graph, nodes1, nodes2=None, assume_disjoint=False, assume_dense=True)[source]¶
Get edges between two components or within a single component
- Parameters:
graph (nx.Graph) – the graph
nodes1 (set) – list of nodes
nodes2 (set) – if None it is equivlanet to nodes2=nodes1 (default=None)
assume_disjoint (bool) – skips expensive check to ensure edges arnt returned twice (default=False)
Example
>>> edges = [ >>> (1, 2), (2, 3), (3, 4), (4, 1), (4, 3), # cc 1234 >>> (1, 5), (7, 2), (5, 1), # cc 567 / 5678 >>> (7, 5), (5, 6), (8, 7), >>> ] >>> digraph = nx.DiGraph(edges) >>> graph = nx.Graph(edges) >>> nodes1 = [1, 2, 3, 4] >>> nodes2 = [5, 6, 7] >>> n2 = sorted(edges_between(graph, nodes1, nodes2)) >>> n4 = sorted(edges_between(graph, nodes1)) >>> n5 = sorted(edges_between(graph, nodes1, nodes1)) >>> n1 = sorted(edges_between(digraph, nodes1, nodes2)) >>> n3 = sorted(edges_between(digraph, nodes1)) >>> print('n2 == %r' % (n2,)) >>> print('n4 == %r' % (n4,)) >>> print('n5 == %r' % (n5,)) >>> print('n1 == %r' % (n1,)) >>> print('n3 == %r' % (n3,)) >>> assert n2 == ([(1, 5), (2, 7)]), '2' >>> assert n4 == ([(1, 2), (1, 4), (2, 3), (3, 4)]), '4' >>> assert n5 == ([(1, 2), (1, 4), (2, 3), (3, 4)]), '5' >>> assert n1 == ([(1, 5), (5, 1), (7, 2)]), '1' >>> assert n3 == ([(1, 2), (2, 3), (3, 4), (4, 1), (4, 3)]), '3' >>> n6 = sorted(edges_between(digraph, nodes1 + [6], nodes2 + [1, 2], assume_dense=False)) >>> print('n6 = %r' % (n6,)) >>> n6 = sorted(edges_between(digraph, nodes1 + [6], nodes2 + [1, 2], assume_dense=True)) >>> print('n6 = %r' % (n6,)) >>> assert n6 == ([(1, 2), (1, 5), (2, 3), (4, 1), (5, 1), (5, 6), (7, 2)]), '6'
- graphid.util.edges_cross(graph, nodes1, nodes2)[source]¶
Finds edges between two sets of disjoint nodes. Running time is O(len(nodes1) * len(nodes2))
- Parameters:
graph (nx.Graph) – an undirected graph
nodes1 (set) – set of nodes disjoint from nodes2
nodes2 (set) – set of nodes disjoint from nodes1.
- graphid.util.edges_inside(graph, nodes)[source]¶
Finds edges within a set of nodes Running time is O(len(nodes) ** 2)
- Parameters:
graph (nx.Graph) – an undirected graph
nodes1 (set) – a set of nodes
- graphid.util.edges_outgoing(graph, nodes)[source]¶
Finds edges leaving a set of nodes. Average running time is O(len(nodes) * ave_degree(nodes)) Worst case running time is O(G.number_of_edges()).
- Parameters:
graph (nx.Graph) – a graph
nodes (set) – set of nodes
Example
>>> G = demodata_bridge() >>> nodes = {1, 2, 3, 4} >>> outgoing = edges_outgoing(G, nodes) >>> assert outgoing == {(3, 5), (4, 8)}
- graphid.util.list_roll(list_, n)[source]¶
Like numpy.roll for python lists
- Parameters:
list_ (list)
n (int)
- Return type:
References
http://stackoverflow.com/questions/9457832/python-list-rotation
Example
>>> list_ = [1, 2, 3, 4, 5] >>> n = 2 >>> result = list_roll(list_, n) >>> print(result) [4, 5, 1, 2, 3]
- graphid.util.nx_delete_edge_attr(graph, name, edges=None)[source]¶
Removes an attributes from specific edges in the graph
Doctest
>>> G = nx.karate_club_graph() >>> nx.set_edge_attributes(G, name='spam', values='eggs') >>> nx.set_edge_attributes(G, name='foo', values='bar') >>> assert len(nx.get_edge_attributes(G, 'spam')) == 78 >>> assert len(nx.get_edge_attributes(G, 'foo')) == 78 >>> nx_delete_edge_attr(G, ['spam', 'foo'], edges=[(1, 2)]) >>> assert len(nx.get_edge_attributes(G, 'spam')) == 77 >>> assert len(nx.get_edge_attributes(G, 'foo')) == 77 >>> nx_delete_edge_attr(G, ['spam']) >>> assert len(nx.get_edge_attributes(G, 'spam')) == 0 >>> assert len(nx.get_edge_attributes(G, 'foo')) == 77
Doctest
>>> G = nx.MultiGraph() >>> G.add_edges_from([(1, 2), (2, 3), (3, 4), (4, 5), (4, 5), (1, 2)]) >>> nx.set_edge_attributes(G, name='spam', values='eggs') >>> nx.set_edge_attributes(G, name='foo', values='bar') >>> assert len(nx.get_edge_attributes(G, 'spam')) == 6 >>> assert len(nx.get_edge_attributes(G, 'foo')) == 6 >>> nx_delete_edge_attr(G, ['spam', 'foo'], edges=[(1, 2, 0)]) >>> assert len(nx.get_edge_attributes(G, 'spam')) == 5 >>> assert len(nx.get_edge_attributes(G, 'foo')) == 5 >>> nx_delete_edge_attr(G, ['spam']) >>> assert len(nx.get_edge_attributes(G, 'spam')) == 0 >>> assert len(nx.get_edge_attributes(G, 'foo')) == 5
- graphid.util.nx_delete_node_attr(graph, name, nodes=None)[source]¶
Removes node attributes
Doctest
>>> G = nx.karate_club_graph() >>> nx.set_node_attributes(G, name='foo', values='bar') >>> datas = nx.get_node_attributes(G, 'club') >>> assert len(nx.get_node_attributes(G, 'club')) == 34 >>> assert len(nx.get_node_attributes(G, 'foo')) == 34 >>> nx_delete_node_attr(G, ['club', 'foo'], nodes=[1, 2]) >>> assert len(nx.get_node_attributes(G, 'club')) == 32 >>> assert len(nx.get_node_attributes(G, 'foo')) == 32 >>> nx_delete_node_attr(G, ['club']) >>> assert len(nx.get_node_attributes(G, 'club')) == 0 >>> assert len(nx.get_node_attributes(G, 'foo')) == 32
- graphid.util.nx_gen_edge_attrs(G, key, edges=None, default=NoParam, on_missing='error', on_keyerr='default')[source]¶
Improved generator version of nx.get_edge_attributes
- Parameters:
on_missing (str) – Strategy for handling nodes missing from G. Can be {‘error’, ‘default’, ‘filter’}. defaults to ‘error’. is on_missing is not error, then we allow any edge even if the endpoints are not in the graph.
on_keyerr (str) – Strategy for handling keys missing from node dicts. Can be {‘error’, ‘default’, ‘filter’}. defaults to ‘default’ if default is specified, otherwise defaults to ‘error’.
CommandLine
python -m graphid.util.nx_utils nx_gen_edge_attrs
Example
>>> from graphid import util >>> from functools import partial >>> G = nx.Graph([(1, 2), (2, 3), (3, 4)]) >>> nx.set_edge_attributes(G, name='part', values={(1, 2): 'bar', (2, 3): 'baz'}) >>> edges = [(1, 2), (2, 3), (3, 4), (4, 5)] >>> func = partial(nx_gen_edge_attrs, G, 'part', default=None) >>> # >>> assert len(list(func(on_missing='error', on_keyerr='default'))) == 3 >>> assert len(list(func(on_missing='error', on_keyerr='filter'))) == 2 >>> util.assert_raises(KeyError, list, func(on_missing='error', on_keyerr='error')) >>> # >>> assert len(list(func(edges, on_missing='filter', on_keyerr='default'))) == 3 >>> assert len(list(func(edges, on_missing='filter', on_keyerr='filter'))) == 2 >>> util.assert_raises(KeyError, list, func(edges, on_missing='filter', on_keyerr='error')) >>> # >>> assert len(list(func(edges, on_missing='default', on_keyerr='default'))) == 4 >>> assert len(list(func(edges, on_missing='default', on_keyerr='filter'))) == 2 >>> util.assert_raises(KeyError, list, func(edges, on_missing='default', on_keyerr='error'))
- graphid.util.nx_gen_edge_values(G, key, edges=None, default=NoParam, on_missing='error', on_keyerr='default')[source]¶
Generates attributes values of specific edges
- Parameters:
on_missing (str) – Strategy for handling nodes missing from G. Can be {‘error’, ‘default’}. defaults to ‘error’.
on_keyerr (str) – Strategy for handling keys missing from node dicts. Can be {‘error’, ‘default’}. defaults to ‘default’ if default is specified, otherwise defaults to ‘error’.
- graphid.util.nx_gen_node_attrs(G, key, nodes=None, default=NoParam, on_missing='error', on_keyerr='default')[source]¶
Improved generator version of nx.get_node_attributes
- Parameters:
on_missing (str) – Strategy for handling nodes missing from G. Can be {‘error’, ‘default’, ‘filter’}. defaults to ‘error’.
on_keyerr (str) – Strategy for handling keys missing from node dicts. Can be {‘error’, ‘default’, ‘filter’}. defaults to ‘default’ if default is specified, otherwise defaults to ‘error’.
Notes
- strategies are:
error - raises an error if key or node does not exist default - returns node, but uses value specified by default filter - skips the node
Example
>>> # ENABLE_DOCTEST >>> from graphid import util >>> G = nx.Graph([(1, 2), (2, 3)]) >>> nx.set_node_attributes(G, name='part', values={1: 'bar', 3: 'baz'}) >>> nodes = [1, 2, 3, 4] >>> # >>> assert len(list(nx_gen_node_attrs(G, 'part', default=None, on_missing='error', on_keyerr='default'))) == 3 >>> assert len(list(nx_gen_node_attrs(G, 'part', default=None, on_missing='error', on_keyerr='filter'))) == 2 >>> assert_raises(KeyError, list, nx_gen_node_attrs(G, 'part', on_missing='error', on_keyerr='error')) >>> # >>> assert len(list(nx_gen_node_attrs(G, 'part', nodes, default=None, on_missing='filter', on_keyerr='default'))) == 3 >>> assert len(list(nx_gen_node_attrs(G, 'part', nodes, default=None, on_missing='filter', on_keyerr='filter'))) == 2 >>> assert_raises(KeyError, list, nx_gen_node_attrs(G, 'part', nodes, on_missing='filter', on_keyerr='error')) >>> # >>> assert len(list(nx_gen_node_attrs(G, 'part', nodes, default=None, on_missing='default', on_keyerr='default'))) == 4 >>> assert len(list(nx_gen_node_attrs(G, 'part', nodes, default=None, on_missing='default', on_keyerr='filter'))) == 2 >>> assert_raises(KeyError, list, nx_gen_node_attrs(G, 'part', nodes, on_missing='default', on_keyerr='error'))
Example
>>> # DISABLE_DOCTEST >>> # ALL CASES >>> from graphid import util >>> G = nx.Graph([(1, 2), (2, 3)]) >>> nx.set_node_attributes(G, name='full', values={1: 'A', 2: 'B', 3: 'C'}) >>> nx.set_node_attributes(G, name='part', values={1: 'bar', 3: 'baz'}) >>> nodes = [1, 2, 3, 4] >>> attrs = dict(nx_gen_node_attrs(G, 'full')) >>> input_grid = { >>> 'nodes': [None, (1, 2, 3, 4)], >>> 'key': ['part', 'full'], >>> 'default': [ub.NoParam, None], >>> } >>> inputs = util.all_dict_combinations(input_grid) >>> kw_grid = { >>> 'on_missing': ['error', 'default', 'filter'], >>> 'on_keyerr': ['error', 'default', 'filter'], >>> } >>> kws = util.all_dict_combinations(kw_grid) >>> for in_ in inputs: >>> for kw in kws: >>> kw2 = ub.dict_union(kw, in_) >>> #print(kw2) >>> on_missing = kw['on_missing'] >>> on_keyerr = kw['on_keyerr'] >>> if on_keyerr == 'default' and in_['default'] is ub.NoParam: >>> on_keyerr = 'error' >>> will_miss = False >>> will_keyerr = False >>> if on_missing == 'error': >>> if in_['key'] == 'part' and in_['nodes'] is not None: >>> will_miss = True >>> if in_['key'] == 'full' and in_['nodes'] is not None: >>> will_miss = True >>> if on_keyerr == 'error': >>> if in_['key'] == 'part': >>> will_keyerr = True >>> if on_missing == 'default': >>> if in_['key'] == 'full' and in_['nodes'] is not None: >>> will_keyerr = True >>> want_error = will_miss or will_keyerr >>> gen = nx_gen_node_attrs(G, **kw2) >>> try: >>> attrs = list(gen) >>> except KeyError: >>> if not want_error: >>> raise AssertionError('should not have errored') >>> else: >>> if want_error: >>> raise AssertionError('should have errored')
- graphid.util.nx_gen_node_values(G, key, nodes, default=NoParam)[source]¶
Generates attributes values of specific nodes
- graphid.util.random_k_edge_connected_graph(size, k, p=0.1, rng=None)[source]¶
Super hacky way of getting a random k-connected graph
Example
>>> from graphid import util >>> size, k, p = 25, 3, .1 >>> rng = util.ensure_rng(0) >>> gs = [] >>> for x in range(4): >>> G = random_k_edge_connected_graph(size, k, p, rng) >>> gs.append(G) >>> # xdoc: +REQUIRES(--show) >>> pnum_ = util.PlotNums(nRows=2, nSubplots=len(gs)) >>> fnum = 1 >>> for g in gs: >>> util.show_nx(g, fnum=fnum, pnum=pnum_())
- graphid.util.take_column(list_, colx)[source]¶
accepts a list of (indexables) and returns a list of indexables can also return a list of list of indexables if colx is a list
- Parameters:
list_ (list) – list of lists
colx (int or list) – index or key in each sublist get item
- Returns:
list of selected items
- Return type:
- Example0:
>>> list_ = [['a', 'b'], ['c', 'd']] >>> colx = 0 >>> result = take_column(list_, colx) >>> result = ub.urepr(result, nl=False) >>> print(result) ['a', 'c']
- Example1:
>>> list_ = [['a', 'b'], ['c', 'd']] >>> colx = [1, 0] >>> result = take_column(list_, colx) >>> result = ub.urepr(result, nl=False) >>> print(result) [['b', 'a'], ['d', 'c']]
- Example2:
>>> list_ = [{'spam': 'EGGS', 'ham': 'SPAM'}, {'spam': 'JAM', 'ham': 'PRAM'}] >>> # colx can be a key or list of keys as well >>> colx = ['spam'] >>> result = take_column(list_, colx) >>> result = ub.urepr(result, nl=False) >>> print(result) [['EGGS'], ['JAM']]
- class graphid.util.PriorityQueue(items=None, ascending=True)[source]¶
Bases:
NiceReprabstracted priority queue for our needs
Combines properties of dicts and heaps Uses a heap for fast minimum/maximum value search Uses a dict for fast read only operations
References
http://code.activestate.com/recipes/522995-priority-dict-a-priority-queue-with-updatable-prio/ https://stackoverflow.com/questions/33024215/built-in-max-heap-api-in-python
Example
>>> items = dict(a=42, b=29, c=40, d=95, e=10) >>> self = PriorityQueue(items) >>> print(self) >>> assert len(self) == 5 >>> print(self.pop()) >>> assert len(self) == 4 >>> print(self.pop()) >>> assert len(self) == 3 >>> print(self.pop()) >>> print(self.pop()) >>> print(self.pop()) >>> assert len(self) == 0
Example
>>> items = dict(a=(1.0, (2, 3)), b=(1.0, (1, 2)), c=(.9, (3, 2))) >>> self = PriorityQueue(items)
- class graphid.util.Boxes(data, format='xywh')[source]¶
Bases:
NiceReprConverts boxes between different formats as long as the last dimension contains 4 coordinates and the format is specified.
This is a convinience class, and should not not store the data for very long. The general idiom should be create class, convert data, and then get the raw data and let the class be garbage collected. This will help ensure that your code is portable and understandable if this class is not available.
Example
>>> # xdoctest: +IGNORE_WHITESPACE >>> Boxes([25, 30, 15, 10], 'xywh') <Boxes(xywh, array([25, 30, 15, 10]))> >>> Boxes([25, 30, 15, 10], 'xywh').to_xywh() <Boxes(xywh, array([25, 30, 15, 10]))> >>> Boxes([25, 30, 15, 10], 'xywh').to_cxywh() <Boxes(cxywh, array([32.5, 35. , 15. , 10. ]))> >>> Boxes([25, 30, 15, 10], 'xywh').to_tlbr() <Boxes(tlbr, array([25, 30, 40, 40]))> >>> Boxes([25, 30, 15, 10], 'xywh').scale(2).to_tlbr() <Boxes(tlbr, array([50., 60., 80., 80.]))>
Example
>>> datas = [ >>> [1, 2, 3, 4], >>> [[1, 2, 3, 4], [4, 5, 6, 7]], >>> [[[1, 2, 3, 4], [4, 5, 6, 7]]], >>> ] >>> formats = ['xywh', 'cxywh', 'tlbr'] >>> for format1 in formats: >>> for data in datas: >>> self = box1 = Boxes(data, format1) >>> for format2 in formats: >>> box2 = box1.toformat(format2) >>> back = box2.toformat(format1) >>> assert box1 == back
- classmethod random(num=1, scale=1.0, format='xywh', rng=None)[source]¶
Makes random boxes
Example
>>> # xdoctest: +IGNORE_WHITESPACE >>> Boxes.random(3, rng=0, scale=100) <Boxes(xywh, array([[27, 35, 30, 27], [21, 32, 21, 44], [48, 19, 39, 26]]))>
- scale(factor)[source]¶
works with tlbr, cxywh, xywh, xy, or wh formats
Example
>>> # xdoctest: +IGNORE_WHITESPACE >>> Boxes(np.array([1, 1, 10, 10])).scale(2).data array([ 2., 2., 20., 20.]) >>> Boxes(np.array([[1, 1, 10, 10]])).scale((2, .5)).data array([[ 2. , 0.5, 20. , 5. ]]) >>> Boxes(np.array([[10, 10]])).scale(.5).data array([[5., 5.]])
- shift(amount)[source]¶
Example
>>> # xdoctest: +IGNORE_WHITESPACE >>> Boxes([25, 30, 15, 10], 'xywh').shift(10) <Boxes(xywh, array([35., 40., 15., 10.]))> >>> Boxes([25, 30, 15, 10], 'xywh').shift((10, 0)) <Boxes(xywh, array([35., 30., 15., 10.]))> >>> Boxes([25, 30, 15, 10], 'tlbr').shift((10, 5)) <Boxes(tlbr, array([35., 35., 25., 15.]))>
- property center¶
- property shape¶
- property area¶
- property components¶
- clip(x_min, y_min, x_max, y_max, inplace=False)[source]¶
Clip boxes to image boundaries. If box is in tlbr format, inplace operation is an option.
Example
>>> # xdoctest: +IGNORE_WHITESPACE >>> boxes = Boxes(np.array([[-10, -10, 120, 120], [1, -2, 30, 50]]), 'tlbr') >>> clipped = boxes.clip(0, 0, 110, 100, inplace=False) >>> assert np.any(boxes.data != clipped.data) >>> clipped2 = boxes.clip(0, 0, 110, 100, inplace=True) >>> assert clipped2.data is boxes.data >>> assert np.all(clipped2.data == clipped.data) >>> print(clipped) <Boxes(tlbr, array([[ 0, 0, 110, 100], [ 1, 0, 30, 50]]))>
- graphid.util.box_ious_py(boxes1, boxes2, bias=1)[source]¶
This is the fastest python implementation of bbox_ious I found
- graphid.util.grab_test_imgpath(key='astro.png', allow_external=True, verbose=True)[source]¶
Gets paths to standard / fun test images. Downloads them if they dont exits
- Parameters:
key (str) – one of the standard test images, e.g. astro.png, carl.jpg, …
allow_external (bool) – if True you can specify existing fpaths
- Returns:
testimg_fpath - filepath to the downloaded or cached test image.
- Return type:
Example
>>> testimg_fpath = grab_test_imgpath('carl.jpg') >>> assert exists(testimg_fpath)
- class graphid.util.GRAPHVIZ_KEYS[source]¶
Bases:
object- N = {'URL', 'area', 'color', 'colorscheme', 'comment', 'distortion', 'fillcolor', 'fixedsize', 'fontcolor', 'fontname', 'fontsize', 'gradientangle', 'group', 'height', 'href', 'id', 'image', 'imagepos', 'imagescale', 'label', 'labelloc', 'layer', 'margin', 'nojustify', 'ordering', 'orientation', 'penwidth', 'peripheries', 'pin', 'pos', 'rects', 'regular', 'root', 'samplepoints', 'shape', 'shapefile', 'showboxes', 'sides', 'skew', 'sortv', 'style', 'target', 'tooltip', 'vertices', 'width', 'xlabel', 'xlp', 'z'}¶
- E = {'URL', 'arrowhead', 'arrowsize', 'arrowtail', 'color', 'colorscheme', 'comment', 'constraint', 'decorate', 'dir', 'edgeURL', 'edgehref', 'edgetarget', 'edgetooltip', 'fillcolor', 'fontcolor', 'fontname', 'fontsize', 'headURL', 'head_lp', 'headclip', 'headhref', 'headlabel', 'headport', 'headtarget', 'headtooltip', 'href', 'id', 'label', 'labelURL', 'labelangle', 'labeldistance', 'labelfloat', 'labelfontcolor', 'labelfontname', 'labelfontsize', 'labelhref', 'labeltarget', 'labeltooltip', 'layer', 'len', 'lhead', 'lp', 'ltail', 'minlen', 'nojustify', 'penwidth', 'pos', 'samehead', 'sametail', 'showboxes', 'style', 'tailURL', 'tail_lp', 'tailclip', 'tailhref', 'taillabel', 'tailport', 'tailtarget', 'tailtooltip', 'target', 'tooltip', 'weight', 'xlabel', 'xlp'}¶
- G = {'Damping', 'K', 'URL', '_background', 'bb', 'bgcolor', 'center', 'charset', 'clusterrank', 'colorscheme', 'comment', 'compound', 'concentrate', 'defaultdist', 'dim', 'dimen', 'diredgeconstraints', 'dpi', 'epsilon', 'esep', 'fontcolor', 'fontname', 'fontnames', 'fontpath', 'fontsize', 'forcelabels', 'gradientangle', 'href', 'id', 'imagepath', 'inputscale', 'label', 'label_scheme', 'labeljust', 'labelloc', 'landscape', 'layerlistsep', 'layers', 'layerselect', 'layersep', 'layout', 'levels', 'levelsgap', 'lheight', 'lp', 'lwidth', 'margin', 'maxiter', 'mclimit', 'mindist', 'mode', 'model', 'mosek', 'newrank', 'nodesep', 'nojustify', 'normalize', 'notranslate', 'nslimit\nnslimit1', 'ordering', 'orientation', 'outputorder', 'overlap', 'overlap_scaling', 'overlap_shrink', 'pack', 'packmode', 'pad', 'page', 'pagedir', 'quadtree', 'quantum', 'rankdir', 'ranksep', 'ratio', 'remincross', 'repulsiveforce', 'resolution', 'root', 'rotate', 'rotation', 'scale', 'searchsize', 'sep', 'showboxes', 'size', 'smoothing', 'sortv', 'splines', 'start', 'style', 'stylesheet', 'target', 'truecolor', 'viewport', 'voro_margin', 'xdotversion'}¶
- graphid.util.bbox_from_extent(extent)[source]¶
- Parameters:
extent (ndarray) – tl_x, br_x, tl_y, br_y
- Returns:
tl_x, tl_y, w, h
- Return type:
bbox (ndarray)
Example
>>> extent = [0, 10, 0, 10] >>> bbox = bbox_from_extent(extent) >>> print('bbox = {}'.format(ub.urepr(list(bbox), nl=0))) bbox = [0, 0, 10, 10]
- graphid.util.draw_network2(graph, layout_info, ax, as_directed=None, hacknoedge=False, hacknode=False, verbose=None, **kwargs)[source]¶
- Kwargs:
use_image, arrow_width, fontsize, fontweight, fontname, fontfamilty, fontproperties
fancy way to draw networkx graphs without directly using networkx
- graphid.util.get_graph_bounding_box(graph)[source]¶
Example
>>> # xdoctest: +REQUIRES(module:pygraphviz) >>> graph = nx.path_graph([1, 2, 3, 4]) >>> nx_agraph_layout(graph, inplace=True) >>> bbox = get_graph_bounding_box(graph) >>> print(ub.urepr(bbox, nl=0)) [0.0, 0.0, 54.0, 252.0]
- graphid.util.netx_draw_images_at_positions(img_list, pos_list, size_list, color_list, framewidth_list)[source]¶
Overlays images on a networkx graph
References
https://gist.github.com/shobhit/3236373 http://matplotlib.org/examples/pylab_examples/demo_annotation_box.html http://stackoverflow.com/questions/11487797/mpl-overlay-small-image http://matplotlib.org/api/text_api.html http://matplotlib.org/api/offsetbox_api.html
- graphid.util.nx_agraph_layout(orig_graph, inplace=False, verbose=None, return_agraph=False, groupby=None, **layoutkw)[source]¶
Uses graphviz and custom code to determine position attributes of nodes and edges.
- Parameters:
groupby (str) – if not None then nodes will be grouped by this attributes and groups will be layed out separately and then stacked together in a grid
References
http://www.graphviz.org/content/attrs http://www.graphviz.org/doc/info/attrs.html
CommandLine
python -m graphid.util.util_graphviz nx_agraph_layout --show
Doctest
>>> # xdoctest: +REQUIRES(module:pygraphviz) >>> from graphid.util.util_graphviz import * # NOQA >>> import networkx as nx >>> import itertools as it >>> from graphid import util >>> n, s = 9, 4 >>> offsets = list(range(0, (1 + n) * s, s)) >>> node_groups = [list(map(str, range(*o))) for o in ub.iter_window(offsets, 2)] >>> edge_groups = [it.combinations(nodes, 2) for nodes in node_groups] >>> graph = nx.Graph() >>> [graph.add_nodes_from(nodes) for nodes in node_groups] >>> [graph.add_edges_from(edges) for edges in edge_groups] >>> for count, nodes in enumerate(node_groups): ... nx.set_node_attributes(graph, name='id', values=ub.dzip(nodes, [count])) >>> layoutkw = dict(prog='neato') >>> graph1, info1 = nx_agraph_layout(graph.copy(), inplace=True, groupby='id', **layoutkw) >>> graph2, info2 = nx_agraph_layout(graph.copy(), inplace=True, **layoutkw) >>> graph3, _ = nx_agraph_layout(graph1.copy(), inplace=True, **layoutkw) >>> nx.set_node_attributes(graph1, name='pin', values='true') >>> graph4, _ = nx_agraph_layout(graph1.copy(), inplace=True, **layoutkw) >>> # xdoc: +REQUIRES(--show) >>> util.show_nx(graph1, layout='custom', pnum=(2, 2, 1), fnum=1) >>> util.show_nx(graph2, layout='custom', pnum=(2, 2, 2), fnum=1) >>> util.show_nx(graph3, layout='custom', pnum=(2, 2, 3), fnum=1) >>> util.show_nx(graph4, layout='custom', pnum=(2, 2, 4), fnum=1) >>> util.show_if_requested() >>> g1pos = nx.get_node_attributes(graph1, 'pos')['1'] >>> g4pos = nx.get_node_attributes(graph4, 'pos')['1'] >>> g2pos = nx.get_node_attributes(graph2, 'pos')['1'] >>> g3pos = nx.get_node_attributes(graph3, 'pos')['1'] >>> print('g1pos = {!r}'.format(g1pos)) >>> print('g4pos = {!r}'.format(g4pos)) >>> print('g2pos = {!r}'.format(g2pos)) >>> print('g3pos = {!r}'.format(g3pos)) >>> assert np.all(g1pos == g4pos), 'points between 1 and 4 were pinned so they should be equal' >>> #assert np.all(g2pos != g3pos), 'points between 2 and 3 were not pinned, so they should be different'
assert np.all(nx.get_node_attributes(graph1, ‘pos’)[‘1’] == nx.get_node_attributes(graph4, ‘pos’)[‘1’]) assert np.all(nx.get_node_attributes(graph2, ‘pos’)[‘1’] == nx.get_node_attributes(graph3, ‘pos’)[‘1’])
- graphid.util.show_nx(graph, with_labels=True, fnum=None, pnum=None, layout='agraph', ax=None, pos=None, img_dict=None, title=None, layoutkw=None, verbose=None, **kwargs)[source]¶
- Parameters:
graph (networkx.Graph)
with_labels (bool) – (default = True)
fnum (int) – figure number(default = None)
pnum (tuple) – plot number(default = None)
layout (str) – (default = ‘agraph’)
ax (None) – (default = None)
pos (None) – (default = None)
img_dict (dict) – (default = None)
title (str) – (default = None)
layoutkw (None) – (default = None)
verbose (bool) – verbosity flag(default = None)
- Kwargs:
use_image, framewidth, modify_ax, as_directed, hacknoedge, hacknode, arrow_width, fontsize, fontweight, fontname, fontfamilty, fontproperties
CommandLine
python -m graphid.util.util_graphviz show_nx --show
Example
>>> # xdoctest: +REQUIRES(module:pygraphviz) >>> from graphid.util.util_graphviz import * # NOQA >>> graph = nx.DiGraph() >>> graph.add_nodes_from(['a', 'b', 'c', 'd']) >>> graph.add_edges_from({'a': 'b', 'b': 'c', 'b': 'd', 'c': 'd'}.items()) >>> nx.set_node_attributes(graph, name='shape', values='rect') >>> nx.set_node_attributes(graph, name='image', values={'a': util.grab_test_imgpath('carl.jpg')}) >>> nx.set_node_attributes(graph, name='image', values={'d': util.grab_test_imgpath('astro.png')}) >>> #nx.set_node_attributes(graph, name='height', values=100) >>> with_labels = True >>> fnum = None >>> pnum = None >>> e = show_nx(graph, with_labels, fnum, pnum, layout='agraph') >>> util.show_if_requested()
- graphid.util.group_pairs(pair_list)[source]¶
Groups a list of items using the first element in each pair as the item and the second element as the groupid.
- Parameters:
pair_list (list) – list of 2-tuples (item, groupid)
- Returns:
groupid_to_items: maps a groupid to a list of items
- Return type:
- graphid.util.grouping_delta(old, new, pure=True)[source]¶
Finds what happened to the old groups to form the new groups.
- Parameters:
old (set of frozensets) – old grouping
new (set of frozensets) – new grouping
pure (bool) – hybrids are separated from pure merges and splits if pure is True, otherwise hybrid cases are grouped in merges and splits.
- Returns:
- delta: dictionary of changes containing the merges, splits,
unchanged, and hybrid cases. Except for unchanged, case a subdict with new and old keys. For splits / merges, one of these contains nested sequences to indicate what the split / merge is. Also reports elements added and removed between old and new if the flattened sets are not the same.
- Return type:
Notes
merges - which old groups were merged into a single new group. splits - which old groups were split into multiple new groups. hybrid - which old groups had split/merge actions applied. unchanged - which old groups are the same as new groups.
Example
>>> # xdoc: +IGNORE_WHITESPACE >>> old = [ >>> [20, 21, 22, 23], [1, 2], [12], [13, 14], [3, 4], [5, 6,11], >>> [7], [8, 9], [10], [31, 32], [33, 34, 35], [41, 42, 43, 44, 45] >>> ] >>> new = [ >>> [20, 21], [22, 23], [1, 2], [12, 13, 14], [4], [5, 6, 3], [7, 8], >>> [9, 10, 11], [31, 32, 33, 34, 35], [41, 42, 43, 44], [45], >>> ] >>> delta = grouping_delta(old, new) >>> assert set(old[0]) in delta['splits']['old'] >>> assert set(new[3]) in delta['merges']['new'] >>> assert set(old[1]) in delta['unchanged'] >>> result = ub.urepr(delta, nl=2, sort=True, nobr=1, sk=True) >>> print(result) hybrid: { merges: [{{10}, {11}, {9}}, {{3}, {5, 6}}, {{4}}, {{7}, {8}}], new: {{3, 5, 6}, {4}, {7, 8}, {9, 10, 11}}, old: {{10}, {3, 4}, {5, 6, 11}, {7}, {8, 9}}, splits: [{{10}}, {{11}, {5, 6}}, {{3}, {4}}, {{7}}, {{8}, {9}}], }, items: { added: {}, removed: {}, }, merges: { new: [{12, 13, 14}, {31, 32, 33, 34, 35}], old: [{{12}, {13, 14}}, {{31, 32}, {33, 34, 35}}], }, splits: { new: [{{20, 21}, {22, 23}}, {{41, 42, 43, 44}, {45}}], old: [{20, 21, 22, 23}, {41, 42, 43, 44, 45}], }, unchanged: { {1, 2}, },
Example
>>> old = [ >>> [1, 2, 3], [4], [5, 6, 7, 8, 9], [10, 11, 12] >>> ] >>> new = [ >>> [1], [2], [3, 4], [5, 6, 7], [8, 9, 10, 11, 12] >>> ] >>> # every case here is hybrid >>> pure_delta = grouping_delta(old, new, pure=True) >>> assert len(list(ub.flatten(pure_delta['merges'].values()))) == 0 >>> assert len(list(ub.flatten(pure_delta['splits'].values()))) == 0 >>> delta = grouping_delta(old, new, pure=False) >>> delta = order_dict_by(delta, ['unchanged', 'splits', 'merges']) >>> result = ub.urepr(delta, nl=2, sort=True, sk=True) >>> print(result) { items: { added: {}, removed: {}, }, merges: [ [{3}, {4}], [{10, 11, 12}, {8, 9}], ], splits: [ [{1}, {2}, {3}], [{5, 6, 7}, {8, 9}], ], unchanged: {}, }
Example
>>> delta = grouping_delta([[1, 2, 3]], []) >>> assert len(delta['items']['removed']) == 3 >>> delta = grouping_delta([], [[1, 2, 3]]) >>> assert len(delta['items']['added']) == 3 >>> delta = grouping_delta([[1]], [[1, 2, 3]]) >>> assert len(delta['items']['added']) == 2 >>> assert len(delta['unchanged']) == 1
- graphid.util.order_dict_by(dict_, key_order)[source]¶
Reorders items in a dictionary according to a custom key order
- Parameters:
dict_ (dict_) – a dictionary
key_order (list) – custom key order
- Returns:
sorted_dict
- Return type:
OrderedDict
Example
>>> dict_ = {1: 1, 2: 2, 3: 3, 4: 4} >>> key_order = [4, 2, 3, 1] >>> sorted_dict = order_dict_by(dict_, key_order) >>> result = ('sorted_dict = %s' % (ub.urepr(sorted_dict, nl=False),)) >>> print(result) >>> assert result == 'sorted_dict = {4: 4, 2: 2, 3: 3, 1: 1}'
- graphid.util.sort_dict(dict_, part='keys', key=None, reverse=False)[source]¶
sorts a dictionary by its values or its keys
- Parameters:
dict_ (dict_) – a dictionary
part (str) – specifies to sort by keys or values
key (Optional[func]) – a function that takes specified part and returns a sortable value
reverse (bool) – (Defaults to False) - True for descinding order. False for ascending order.
- Returns:
sorted dictionary
- Return type:
OrderedDict
Example
>>> dict_ = {'a': 3, 'c': 2, 'b': 1} >>> results = [] >>> results.append(sort_dict(dict_, 'keys')) >>> results.append(sort_dict(dict_, 'vals')) >>> results.append(sort_dict(dict_, 'vals', lambda x: -x)) >>> result = ub.urepr(results) >>> print(result) [ {'a': 3, 'b': 1, 'c': 2}, {'b': 1, 'c': 2, 'a': 3}, {'a': 3, 'c': 2, 'b': 1}, ]
- graphid.util.sortedby(item_list, key_list, reverse=False)[source]¶
sorts
item_listusing key_list- Parameters:
list_ (list) – list to sort
key_list (list) – list to sort by
reverse (bool) – sort order is descending (largest first) if reverse is True else acscending (smallest first)
- Returns:
list_sorted by the values of anotherlist. defaults to ascending order- Return type:
- SeeAlso:
sortedby2
Examples
>>> list_ = [1, 2, 3, 4, 5] >>> key_list = [2, 5, 3, 1, 5] >>> result = sortedby(list_, key_list, reverse=True) >>> print(result) [5, 2, 3, 1, 4]
- graphid.util.convert_colorspace(img, dst_space, src_space='BGR', copy=False, dst=None)[source]¶
Converts colorspace of img. Convinience function around cv2.cvtColor
- Parameters:
img (ndarray[uint8_t, ndim=2]) – image data
colorspace (str) – RGB, LAB, etc
dst_space (unicode) – (default = u’BGR’)
- Returns:
img - image data
- Return type:
ndarray[uint8_t, ndim=2]
Example
>>> convert_colorspace(np.array([[[0, 0, 1]]], dtype=np.float32), 'LAB', src_space='RGB') >>> convert_colorspace(np.array([[[0, 1, 0]]], dtype=np.float32), 'LAB', src_space='RGB') >>> convert_colorspace(np.array([[[1, 0, 0]]], dtype=np.float32), 'LAB', src_space='RGB') >>> convert_colorspace(np.array([[[1, 1, 1]]], dtype=np.float32), 'LAB', src_space='RGB') >>> convert_colorspace(np.array([[[0, 0, 1]]], dtype=np.float32), 'HSV', src_space='RGB')
- graphid.util.ensure_float01(img, dtype=<class 'numpy.float32'>, copy=True)[source]¶
Ensure that an image is encoded using a float properly
- graphid.util.imread(fpath, **kw)[source]¶
reads image data in BGR format
Example
>>> # xdoctest: +SKIP("use kwimage.imread") >>> import ubelt as ub >>> import tempfile >>> from os.path import splitext # NOQA >>> fpath = ub.grabdata('https://i.imgur.com/oHGsmvF.png', fname='carl.png') >>> #fpath = ub.grabdata('http://www.topcoder.com/contest/problem/UrbanMapper3D/JAX_Tile_043_DTM.tif') >>> ext = splitext(fpath)[1] >>> img1 = imread(fpath) >>> # Check that write + read preserves data >>> tmp = tempfile.NamedTemporaryFile(suffix=ext) >>> imwrite(tmp.name, img1) >>> img2 = imread(tmp.name) >>> assert np.all(img2 == img1)
Example
>>> # xdoctest: +SKIP("use kwimage.imread") >>> import tempfile >>> import ubelt as ub >>> #img1 = (np.arange(0, 12 * 12 * 3).reshape(12, 12, 3) % 255).astype(np.uint8) >>> img1 = imread(ub.grabdata('http://i.imgur.com/iXNf4Me.png', fname='ada.png')) >>> tmp_tif = tempfile.NamedTemporaryFile(suffix='.tif') >>> tmp_png = tempfile.NamedTemporaryFile(suffix='.png') >>> imwrite(tmp_tif.name, img1) >>> imwrite(tmp_png.name, img1) >>> tif_im = imread(tmp_tif.name) >>> png_im = imread(tmp_png.name) >>> assert np.all(tif_im == png_im)
Example
>>> # xdoctest: +SKIP("use kwimage.imread") >>> from graphid.util.util_image import * >>> import tempfile >>> import ubelt as ub >>> #img1 = (np.arange(0, 12 * 12 * 3).reshape(12, 12, 3) % 255).astype(np.uint8) >>> tif_fpath = ub.grabdata('https://ghostscript.com/doc/tiff/test/images/rgb-3c-16b.tiff') >>> img1 = imread(tif_fpath) >>> tmp_tif = tempfile.NamedTemporaryFile(suffix='.tif') >>> tmp_png = tempfile.NamedTemporaryFile(suffix='.png') >>> imwrite(tmp_tif.name, img1) >>> imwrite(tmp_png.name, img1) >>> tif_im = imread(tmp_tif.name) >>> png_im = imread(tmp_png.name) >>> assert np.all(tif_im == png_im)
- class graphid.util.KWSpec(spec)[source]¶
Bases:
objectSafer keyword arguments with keyword specifications.
- graphid.util.all_dict_combinations(varied_dict)[source]¶
- Parameters:
varied_dict (dict) – a dict with lists of possible parameter settings
- Returns:
dict_list a list of dicts correpsonding to all combinations of params settings
- Return type:
Example
>>> varied_dict = {'logdist_weight': [0.0, 1.0], 'pipeline_root': ['vsmany'], 'sv_on': [True, False, None]} >>> dict_list = all_dict_combinations(varied_dict) >>> result = str(ub.urepr(dict_list)) >>> print(result) [ {'logdist_weight': 0.0, 'pipeline_root': 'vsmany', 'sv_on': True}, {'logdist_weight': 0.0, 'pipeline_root': 'vsmany', 'sv_on': False}, {'logdist_weight': 0.0, 'pipeline_root': 'vsmany', 'sv_on': None}, {'logdist_weight': 1.0, 'pipeline_root': 'vsmany', 'sv_on': True}, {'logdist_weight': 1.0, 'pipeline_root': 'vsmany', 'sv_on': False}, {'logdist_weight': 1.0, 'pipeline_root': 'vsmany', 'sv_on': None}, ]
- graphid.util.aslist(sequence)[source]¶
Ensures that the sequence object is a Python list. Handles, numpy arrays, and python sequences (e.g. tuples, and iterables).
- Parameters:
sequence (sequence) – a list-like object
- Returns:
list_ - sequence as a Python list
- Return type:
Example
>>> s1 = [1, 2, 3] >>> s2 = (1, 2, 3) >>> assert aslist(s1) is s1 >>> assert aslist(s2) is not s2 >>> aslist(np.array([[1, 2], [3, 4], [5, 6]])) [[1, 2], [3, 4], [5, 6]] >>> aslist(range(3)) [0, 1, 2]
- class graphid.util.classproperty(fget=None, fset=None, fdel=None, doc=None)[source]¶
Bases:
propertyDecorates a method turning it into a classattribute
References
https://stackoverflow.com/questions/1697501/python-staticmethod-with-property
- graphid.util.cprint(text, color=None)[source]¶
provides some color to terminal output
- Parameters:
text (str)
color (str)
- Example0:
>>> import pygments.console >>> msg_list = list(pygments.console.codes.keys()) >>> color_list = list(pygments.console.codes.keys()) >>> [cprint(text, color) for text, color in zip(msg_list, color_list)]
- Example1:
>>> import pygments.console >>> print('line1') >>> cprint('line2', 'red') >>> cprint('line3', 'blue') >>> cprint('line4', 'magenta') >>> cprint('line5', 'reset') >>> cprint('line5', 'magenta') >>> print('line6')
- graphid.util.delete_dict_keys(dict_, key_list)[source]¶
Removes items from a dictionary inplace. Keys that do not exist are ignored.
- Parameters:
dict_ (dict) – dict like object with a __del__ attribute
key_list (list) – list of keys that specify the items to remove
Example
>>> dict_ = {'bread': 1, 'churches': 1, 'cider': 2, 'very small rocks': 2} >>> key_list = ['duck', 'bread', 'cider'] >>> delete_dict_keys(dict_, key_list) >>> result = ub.urepr(dict_, nl=False) >>> print(result) {'churches': 1, 'very small rocks': 2}
- graphid.util.delete_items_by_index(list_, index_list, copy=False)[source]¶
Remove items from
list_at positions specified inindex_listThe originallist_is preserved ifcopyis True- Parameters:
list_ (list)
index_list (list)
copy (bool) – preserves original list if True
Example
>>> list_ = [8, 1, 8, 1, 6, 6, 3, 4, 4, 5, 6] >>> index_list = [2, -1] >>> result = delete_items_by_index(list_, index_list) >>> print(result) [8, 1, 1, 6, 6, 3, 4, 4, 5]
- graphid.util.ensure_iterable(obj)[source]¶
- Parameters:
obj (scalar or iterable)
- Returns:
obj if it was iterable otherwise [obj]
- Return type:
it3erable
- Timeit:
%timeit util.ensure_iterable([1]) %timeit util.ensure_iterable(1) %timeit util.ensure_iterable(np.array(1)) %timeit util.ensure_iterable([1]) %timeit [1]
Example
>>> obj_list = [3, [3], '3', (3,), [3,4,5]] >>> result = [ensure_iterable(obj) for obj in obj_list] >>> result = str(result) >>> print(result) [[3], [3], ['3'], (3,), [3, 4, 5]]
- graphid.util.estarmap(func, iter_, **kwargs)[source]¶
Eager version of it.starmap from itertools
Note this is inefficient and should only be used when prototyping and debugging.
- graphid.util.get_timestamp(format_='iso', use_second=False, delta_seconds=None, isutc=False, timezone=False)[source]¶
- Parameters:
format_ (str) – (tag, printable, filename, other)
use_second (bool)
delta_seconds (None)
- Returns:
stamp
- Return type:
Example
>>> format_ = 'printable' >>> use_second = False >>> delta_seconds = None >>> stamp = get_timestamp(format_, use_second, delta_seconds) >>> print(stamp) >>> assert len(stamp) == len('15:43:04 2015/02/24')
- graphid.util.isect(list1, list2)[source]¶
returns list1 elements that are also in list2. preserves order of list1
intersect_ordered
- Parameters:
list1 (list)
list2 (list)
- Returns:
new_list
- Return type:
Example
>>> list1 = ['featweight_rowid', 'feature_rowid', 'config_rowid', 'featweight_forground_weight'] >>> list2 = [u'featweight_rowid'] >>> result = isect(list1, list2) >>> print(result) ['featweight_rowid']
- graphid.util.make_index_lookup(list_, dict_factory=<class 'dict'>)[source]¶
- Parameters:
list_ (list) – assumed to have unique items
- Returns:
mapping from item to index
- Return type:
Example
>>> list_ = [5, 3, 8, 2] >>> idx2_item = make_index_lookup(list_) >>> result = ub.urepr(idx2_item, nl=False, sort=1) >>> assert list(ub.take(idx2_item, list_)) == list(range(len(list_))) >>> print(result) {2: 3, 3: 1, 5: 0, 8: 2}
- graphid.util.replace_nones(list_, repl=-1)[source]¶
Recursively removes Nones in all lists and sublists and replaces them with the repl variable
- Parameters:
list_ (list)
repl (obj) – replacement value
- Returns:
list
Example
>>> list_ = [None, 0, 1, 2] >>> repl = -1 >>> repl_list = replace_nones(list_, repl) >>> result = str(repl_list) >>> print(result) [-1, 0, 1, 2]
- graphid.util.safe_argmax(arr, fill=nan, finite=False, nans=True)[source]¶
Doctest
>>> assert safe_argmax([np.nan, np.nan], nans=False) == 0 >>> assert safe_argmax([-100, np.nan], nans=False) == 0 >>> assert safe_argmax([np.nan, -100], nans=False) == 1 >>> assert safe_argmax([-100, 0], nans=False) == 1 >>> assert np.isnan(safe_argmax([]))
- graphid.util.safe_extreme(arr, op, fill=nan, finite=False, nans=True)[source]¶
Applies an exterme operation to an 1d array (typically max/min) but ensures a value is always returned even in operations without identities. The default identity must be specified using the fill argument.
- Parameters:
arr (ndarray) – 1d array to take extreme of
op (func) – vectorized operation like np.max to apply to array
fill (float) – return type if arr has no elements (default = nan)
finite (bool) – if True ignores non-finite values (default = False)
nans (bool) – if False ignores nans (default = True)
- graphid.util.safe_max(arr, fill=nan, finite=False, nans=True)[source]¶
- Parameters:
arr (ndarray) – 1d array to take max of
fill (float) – return type if arr has no elements (default = nan)
finite (bool) – if True ignores non-finite values (default = False)
nans (bool) – if False ignores nans (default = True)
Example
>>> arrs = [[], [np.nan], [-np.inf, np.nan, np.inf], [np.inf], [np.inf, 1], [0, 1]] >>> arrs = [np.array(arr) for arr in arrs] >>> fill = np.nan >>> results1 = [safe_max(arr, fill, finite=False, nans=True) for arr in arrs] >>> results2 = [safe_max(arr, fill, finite=True, nans=True) for arr in arrs] >>> results3 = [safe_max(arr, fill, finite=True, nans=False) for arr in arrs] >>> results4 = [safe_max(arr, fill, finite=False, nans=False) for arr in arrs] >>> results = [results1, results2, results3, results4] >>> result = ('results = %s' % (ub.urepr(results, nl=1, sv=1),)) >>> print(result) results = [ [nan, nan, nan, inf, inf, 1], [nan, nan, nan, nan, 1.0, 1], [nan, nan, nan, nan, 1.0, 1], [nan, nan, inf, inf, inf, 1], ]
- graphid.util.safe_min(arr, fill=nan, finite=False, nans=True)[source]¶
Example
>>> arrs = [[], [np.nan], [-np.inf, np.nan, np.inf], [np.inf], [np.inf, 1], [0, 1]] >>> arrs = [np.array(arr) for arr in arrs] >>> fill = np.nan >>> results1 = [safe_min(arr, fill, finite=False, nans=True) for arr in arrs] >>> results2 = [safe_min(arr, fill, finite=True, nans=True) for arr in arrs] >>> results3 = [safe_min(arr, fill, finite=True, nans=False) for arr in arrs] >>> results4 = [safe_min(arr, fill, finite=False, nans=False) for arr in arrs] >>> results = [results1, results2, results3, results4] >>> result = ('results = %s' % (ub.urepr(results, nl=1, sv=1),)) >>> print(result) results = [ [nan, nan, nan, inf, 1.0, 0], [nan, nan, nan, nan, 1.0, 0], [nan, nan, nan, nan, 1.0, 0], [nan, nan, -inf, inf, 1.0, 0], ]
- graphid.util.setdiff(list1, list2)[source]¶
returns list1 elements that are not in list2. preserves order of list1
- Parameters:
list1 (list)
list2 (list)
- Returns:
new_list
- Return type:
Example
>>> list1 = ['featweight_rowid', 'feature_rowid', 'config_rowid', 'featweight_forground_weight'] >>> list2 = [u'featweight_rowid'] >>> new_list = setdiff(list1, list2) >>> result = ub.urepr(new_list, nl=False) >>> print(result) ['feature_rowid', 'config_rowid', 'featweight_forground_weight']
- graphid.util.snapped_slice(size, frac, n)[source]¶
Creates a slice spanning n items in a list of length size at position frac.
- Parameters:
size (int) – length of the list
frac (float) – position in the range [0, 1]
n (int) – number of items in the slice
- Returns:
slice object that best fits the criteria
- Return type:
- SeeAlso:
take_percentile_parts
Example:
Example
>>> # DISABLE_DOCTEST >>> print(snapped_slice(0, 0, 10)) >>> print(snapped_slice(1, 0, 10)) >>> print(snapped_slice(100, 0, 10)) >>> print(snapped_slice(9, 0, 10)) >>> print(snapped_slice(100, 1, 10)) pass
- graphid.util.stats_dict(list_, axis=None, use_nan=False, use_sum=False, use_median=False, size=False)[source]¶
- Parameters:
list_ (listlike) – values to get statistics of
axis (int) – if list_ is ndarray then this specifies the axis
- Returns:
- stats: dictionary of common numpy statistics
(min, max, mean, std, nMin, nMax, shape)
- Return type:
OrderedDict
- Examples0:
>>> # xdoctest: +IGNORE_WHITESPACE >>> import numpy as np >>> axis = 0 >>> np.random.seed(0) >>> list_ = np.random.rand(10, 2).astype(np.float32) >>> stats = stats_dict(list_, axis, use_nan=False) >>> result = str(ub.urepr(stats, nl=1, precision=4, with_dtype=True)) >>> print(result) { 'mean': np.array([0.5206, 0.6425], dtype=np.float32), 'std': np.array([0.2854, 0.2517], dtype=np.float32), 'max': np.array([0.9637, 0.9256], dtype=np.float32), 'min': np.array([0.0202, 0.0871], dtype=np.float32), 'nMin': np.array([1, 1], dtype=np.int32), 'nMax': np.array([1, 1], dtype=np.int32), 'shape': (10, 2), }
- Examples1:
>>> import numpy as np >>> axis = 0 >>> rng = np.random.RandomState(0) >>> list_ = rng.randint(0, 42, size=100).astype(np.float32) >>> list_[4] = np.nan >>> stats = stats_dict(list_, axis, use_nan=True) >>> result = str(ub.urepr(stats, precision=1, sk=True)) >>> print(result) {mean: 20.0, std: 13.2, max: 41.0, min: 0.0, nMin: 7, nMax: 3, shape: (100,), num_nan: 1,}
- graphid.util.take_percentile_parts(arr, front=None, mid=None, back=None)[source]¶
Take parts from front, back, or middle of a list
Example
>>> arr = list(range(20)) >>> front = 3 >>> mid = 3 >>> back = 3 >>> result = take_percentile_parts(arr, front, mid, back) >>> print(result) [0, 1, 2, 9, 10, 11, 17, 18, 19]
- graphid.util.apply_grouping(items, groupxs, axis=0)[source]¶
applies grouping from group_indicies apply_grouping
- Parameters:
items (ndarray)
groupxs (list of ndarrays)
- Returns:
grouped items
- Return type:
list of ndarrays
- SeeAlso:
group_indices invert_apply_grouping
Example
>>> # xdoctest: +IGNORE_WHITESPACE >>> idx2_groupid = np.array([2, 1, 2, 1, 2, 1, 2, 3, 3, 3, 3]) >>> items = np.array([1, 8, 5, 5, 8, 6, 7, 5, 3, 0, 9]) >>> (keys, groupxs) = group_indices(idx2_groupid) >>> grouped_items = apply_grouping(items, groupxs) >>> result = str(grouped_items) >>> print(result) [array([8, 5, 6]), array([1, 5, 8, 7]), array([5, 3, 0, 9])]
- graphid.util.atleast_nd(arr, n, front=False)[source]¶
View inputs as arrays with at least n dimensions. TODO: Submit as a PR to numpy
- Parameters:
arr (array_like) – One array-like object. Non-array inputs are converted to arrays. Arrays that already have n or more dimensions are preserved.
n (int) – number of dimensions to ensure
tofront (bool) – if True new dimensions are added to the front of the array. otherwise they are added to the back.
- Returns:
An array with
a.ndim >= n. Copies are avoided where possible, and views with three or more dimensions are returned. For example, a 1-D array of shape(N,)becomes a view of shape(1, N, 1), and a 2-D array of shape(M, N)becomes a view of shape(M, N, 1).- Return type:
ndarray
Example
>>> n = 2 >>> arr = np.array([1, 1, 1]) >>> arr_ = atleast_nd(arr, n) >>> result = ub.urepr(arr_.tolist(), nl=0) >>> print(result) [[1], [1], [1]]
Example
>>> n = 4 >>> arr1 = [1, 1, 1] >>> arr2 = np.array(0) >>> arr3 = np.array([[[[[1]]]]]) >>> arr1_ = atleast_nd(arr1, n) >>> arr2_ = atleast_nd(arr2, n) >>> arr3_ = atleast_nd(arr3, n) >>> result1 = ub.urepr(arr1_.tolist(), nl=0) >>> result2 = ub.urepr(arr2_.tolist(), nl=0) >>> result3 = ub.urepr(arr3_.tolist(), nl=0) >>> result = '\n'.join([result1, result2, result3]) >>> print(result) [[[[1]]], [[[1]]], [[[1]]]] [[[[0]]]] [[[[[1]]]]]
Benchmark
import ubelt N = 100
t1 = ubelt.Timerit(N, label=’mine’) for timer in t1:
arr = np.empty((10, 10)) with timer:
atleast_nd(arr, 3)
t2 = ubelt.Timerit(N, label=’baseline’) for timer in t2:
arr = np.empty((10, 10)) with timer:
np.atleast_3d(arr)
- graphid.util.group_indices(idx2_groupid, assume_sorted=False)[source]¶
- Parameters:
idx2_groupid (ndarray) – numpy array of group ids (must be numeric)
- Returns:
(keys, groupxs)
- Return type:
- Example0:
>>> # xdoctest: +IGNORE_WHITESPACE >>> idx2_groupid = np.array([2, 1, 2, 1, 2, 1, 2, 3, 3, 3, 3]) >>> (keys, groupxs) = group_indices(idx2_groupid) >>> result = ub.urepr((keys, groupxs), nobr=True, with_dtype=True) >>> print(result)
np.array([1, 2, 3], dtype=np.int64), [
np.array([1, 3, 5], dtype=np.int64), np.array([0, 2, 4, 6], dtype=np.int64), np.array([ 7, 8, 9, 10], dtype=np.int64)…
- Example1:
>>> # xdoctest: +IGNORE_WHITESPACE >>> idx2_groupid = np.array([[ 24], [ 129], [ 659], [ 659], [ 24], ... [659], [ 659], [ 822], [ 659], [ 659], [24]]) >>> # 2d arrays must be flattened before coming into this function so >>> # information is on the last axis >>> (keys, groupxs) = group_indices(idx2_groupid.T[0]) >>> result = ub.urepr((keys, groupxs), nobr=True, with_dtype=True) >>> print(result)
np.array([ 24, 129, 659, 822], dtype=np.int64), [
np.array([ 0, 4, 10], dtype=np.int64), np.array([1], dtype=np.int64), np.array([2, 3, 5, 6, 8, 9], dtype=np.int64), np.array([7], dtype=np.int64)…
- Example2:
>>> # xdoctest: +IGNORE_WHITESPACE >>> idx2_groupid = np.array([True, True, False, True, False, False, True]) >>> (keys, groupxs) = group_indices(idx2_groupid) >>> result = ub.urepr((keys, groupxs), nobr=True, with_dtype=True) >>> print(result)
np.array([False, True], dtype=bool), [
np.array([2, 4, 5], dtype=np.int64), np.array([0, 1, 3, 6], dtype=np.int64)…
- Timeit:
import numba group_indices_numba = numba.jit(group_indices) group_indices_numba(idx2_groupid)
- SeeAlso:
apply_grouping
References
http://stackoverflow.com/questions/4651683/ numpy-grouping-using-itertools-groupby-performance
Todo
Look into np.split http://stackoverflow.com/questions/21888406/ getting-the-indexes-to-the-duplicate-columns-of-a-numpy-array
- graphid.util.isect_flags(arr, other)[source]¶
Example
>>> arr = np.array([ >>> [1, 2, 3, 4], >>> [5, 6, 3, 4], >>> [1, 1, 3, 4], >>> ]) >>> other = np.array([1, 4, 6]) >>> mask = isect_flags(arr, other) >>> print(mask) [[ True False False True] [False True False True] [ True True False True]]
- graphid.util.iter_reduce_ufunc(ufunc, arr_iter, out=None)[source]¶
constant memory iteration and reduction
applys ufunc from left to right over the input arrays
Example
>>> arr_list = [ ... np.array([0, 1, 2, 3, 8, 9]), ... np.array([4, 1, 2, 3, 4, 5]), ... np.array([0, 5, 2, 3, 4, 5]), ... np.array([1, 1, 6, 3, 4, 5]), ... np.array([0, 1, 2, 7, 4, 5]) ... ] >>> memory = np.array([9, 9, 9, 9, 9, 9]) >>> gen_memory = memory.copy() >>> def arr_gen(arr_list, gen_memory): ... for arr in arr_list: ... gen_memory[:] = arr ... yield gen_memory >>> print('memory = %r' % (memory,)) >>> print('gen_memory = %r' % (gen_memory,)) >>> ufunc = np.maximum >>> res1 = iter_reduce_ufunc(ufunc, iter(arr_list), out=None) >>> res2 = iter_reduce_ufunc(ufunc, iter(arr_list), out=memory) >>> res3 = iter_reduce_ufunc(ufunc, arr_gen(arr_list, gen_memory), out=memory) >>> print('res1 = %r' % (res1,)) >>> print('res2 = %r' % (res2,)) >>> print('res3 = %r' % (res3,)) >>> print('memory = %r' % (memory,)) >>> print('gen_memory = %r' % (gen_memory,)) >>> assert np.all(res1 == res2) >>> assert np.all(res2 == res3)
- graphid.util.ensure_rng(rng, api='numpy')[source]¶
Returns a random number generator
- Parameters:
seed – if None, then the rng is unseeded. Otherwise the seed can be an integer or a RandomState class
Example
>>> rng = ensure_rng(None) >>> ensure_rng(0).randint(0, 1000) 684 >>> ensure_rng(np.random.RandomState(1)).randint(0, 1000) 37
Example
>>> num = 4 >>> print('--- Python as PYTHON ---') >>> py_rng = random.Random(0) >>> pp_nums = [py_rng.random() for _ in range(num)] >>> print(pp_nums) >>> print('--- Numpy as PYTHON ---') >>> np_rng = ensure_rng(random.Random(0), api='numpy') >>> np_nums = [np_rng.rand() for _ in range(num)] >>> print(np_nums) >>> print('--- Numpy as NUMPY---') >>> np_rng = np.random.RandomState(seed=0) >>> nn_nums = [np_rng.rand() for _ in range(num)] >>> print(nn_nums) >>> print('--- Python as NUMPY---') >>> py_rng = ensure_rng(np.random.RandomState(seed=0), api='python') >>> pn_nums = [py_rng.random() for _ in range(num)] >>> print(pn_nums) >>> assert np_nums == pp_nums >>> assert pn_nums == nn_nums
- graphid.util.random_combinations(items, size, num=None, rng=None)[source]¶
Yields num combinations of length size from items in random order
- Parameters:
items (List) – pool of items to choose from
size (int) – number of items in each combination
num (None, default=None) – number of combinations to generate
rng (int | RandomState, default=None) – seed or random number generator
- Yields:
tuple – combo
Example
>>> import ubelt as ub # NOQA >>> items = list(range(10)) >>> size = 3 >>> num = 5 >>> rng = 0 >>> combos = list(random_combinations(items, size, num, rng)) >>> result = ('combos = %s' % (ub.urepr(combos),)) >>> print(result)
Example
>>> import ubelt as ub # NOQA >>> items = list(zip(range(10), range(10))) >>> size = 3 >>> num = 5 >>> rng = 0 >>> combos = list(random_combinations(items, size, num, rng)) >>> result = ('combos = %s' % (ub.urepr(combos),)) >>> print(result)
- graphid.util.random_product(items, num=None, rng=None)[source]¶
Yields num items from the cartesian product of items in a random order.
- Parameters:
items (list of sequences) – items to get caresian product of packed in a list or tuple. (note this deviates from api of it.product)
Example
>>> items = [(1, 2, 3), (4, 5, 6, 7)] >>> rng = 0 >>> list(random_product(items, rng=0)) >>> list(random_product(items, num=3, rng=0))
- graphid.util.shuffle(items, rng=None)[source]¶
Shuffles a list inplace and then returns it for convinience
- Parameters:
items (list or ndarray) – list to shuffl
rng (RandomState or int) – seed or random number gen
- Returns:
this is the input, but returned for convinience
- Return type:
Example
>>> list1 = [1, 2, 3, 4, 5, 6] >>> list2 = shuffle(list(list1), rng=1) >>> assert list1 != list2 >>> result = str(list2) >>> print(result) [3, 2, 5, 1, 4, 6]
- graphid.util.alias_tags(tags_list, alias_map)[source]¶
update tags to new values
- Parameters:
tags_list (list)
alias_map (list) – list of 2-tuples with regex, value
- Returns:
updated tags
- Return type:
- graphid.util.build_alias_map(regex_map, tag_vocab)[source]¶
Constructs explicit mapping. Order of items in regex map matters. Items at top are given preference.
- graphid.util.filterflags_general_tags(tags_list, has_any=None, has_all=None, has_none=None, min_num=None, max_num=None, any_startswith=None, any_endswith=None, in_any=None, any_match=None, none_match=None, logic='and', ignore_case=True)[source]¶
- Parameters:
tags_list (list)
has_any (None) – (default = None)
has_all (None) – (default = None)
min_num (None) – (default = None)
max_num (None) – (default = None)
Notes
in_any should probably be ni_any
- Example1:
>>> # ENABLE_DOCTEST >>> tags_list = [['v'], [], ['P'], ['P'], ['n', 'o'], [], ['n', 'N'], ['e', 'i', 'p', 'b', 'n'], ['n'], ['n'], ['N']] >>> has_all = 'n' >>> min_num = 1 >>> flags = filterflags_general_tags(tags_list, has_all=has_all, min_num=min_num) >>> result = list(ub.compress(tags_list, flags)) >>> print('result = %r' % (result,))
- Example2:
>>> tags_list = [['vn'], ['vn', 'no'], ['P'], ['P'], ['n', 'o'], [], ['n', 'N'], ['e', 'i', 'p', 'b', 'n'], ['n'], ['n', 'nP'], ['NP']] >>> kwargs = { >>> 'any_endswith': 'n', >>> 'any_match': None, >>> 'any_startswith': 'n', >>> 'has_all': None, >>> 'has_any': None, >>> 'has_none': None, >>> 'max_num': 3, >>> 'min_num': 1, >>> 'none_match': ['P'], >>> } >>> flags = filterflags_general_tags(tags_list, **kwargs) >>> filtered = list(ub.compress(tags_list, flags)) >>> result = ('result = %s' % (ub.urepr(filtered, nl=0),)) >>> print(result) result = [['vn', 'no'], ['n', 'o'], ['n', 'N'], ['n'], ['n', 'nP']]